Our lab explores the relationship between the immune response and gut microbiome in relation to human health and disease. Our current endeavors focus on (1) the development of computational and genomic methods to analyze microbial systems under various conditions, including drug treatments and health states, and (2) the identification of tumoral, systemic, and gut microbial markers that can predict cancer therapy outcomes. Using this information, we will hopefully suggest new ways to prevent, diagnose and treat diseases. We also collaborate on major bioinformatics projects with other researchers like Dr. Rakovitch at Sunnybrook Research Institute on Breast Ductal Carcinoma In Situ, and Prof. Kimmins at Université de Montréal on male infertility and epigenetics.
Overview
Novel approaches to place the molecular properties of cancer within the context of the patient
The majority of cancer research to date has largely focused on understanding molecular properties of the tumor and its surrounding microenvironment in order to therapeutically target key molecular components that might drive disease progression. However, it is now well understood that cancer cells do not exist in isolation. For example, we presented quantitative evidence that the tumor and microenvironment for many breast cancer patients (~20%) do not provide sufficient information to predict how the disease will progress. There does not appear to be any signals at the tumor site that alone are good markers of response to standard of care treatment and ultimately patient prognosis. In turn, this suggests that other factors external to the tumor including, for example, the patient’s immune system, gut microbiome, lifestyle and exposures may play a significant role in disease progression.
In our most recent study, we established the existence of several molecular interactions between the primary tumor and the patient systemic response. That is, we identified molecular processes and pathways in the primary breast tumor that are tightly co-expressed with molecular processes and pathways in the patient blood cells (our surrogate to measure the patient systemic response). Genomic profiling of RNA from a patient’s blood can be used to assess the state of the immune system in an individual and offer many opportunities to participate in precision medicine efforts.
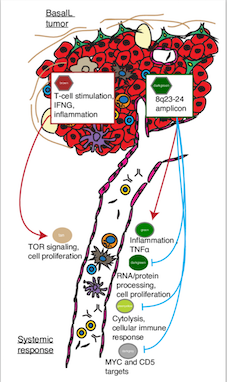
Our efforts to date have harvested RNA from whole blood composed of diverse cell types (eg, T and B lymphocytes, neutrophils, platelets). Thus, molecular profiles of these samples are “averaging” over the transcriptional programs of the different types of cells in blood. Modern approaches based on single-cell RNA sequencing are becoming increasingly routine where RNA from a single cell is isolated and sequenced. This allows transcriptional programs of each type (eg immune) of cell within a blood sample to be measured. Such approaches have great utility when investigating, how an individual is responding to therapy, to the presence/management of specific adverse effects, and ultimately better inform on long term progression of the disease, including the early diagnosis of recurrence.
The gut microbiome has been implicated in a vast number of host’s physiological processes. Analogous to our studies of the immune system, we have also initiated studies to characterize these complex microbial communities, and explore how it is affected by one's lifestyle and how it might benefit the host by impacting functions in another tissue (eg systemic immune response to cancer).
These interactions and additional information regarding patient exposure and life-style information are the first steps towards a new generation of integrative holistic predictors for a broad range of clinical end-points: diagnostics, response to therapies, prognosis and disease monitoring.
Relevant recent papers:
Interactions between the tumor and the blood systemic response of breast cancer patients
The prognostic ease and difficulty of invasive breast carcinoma
Relevant recent software packages:
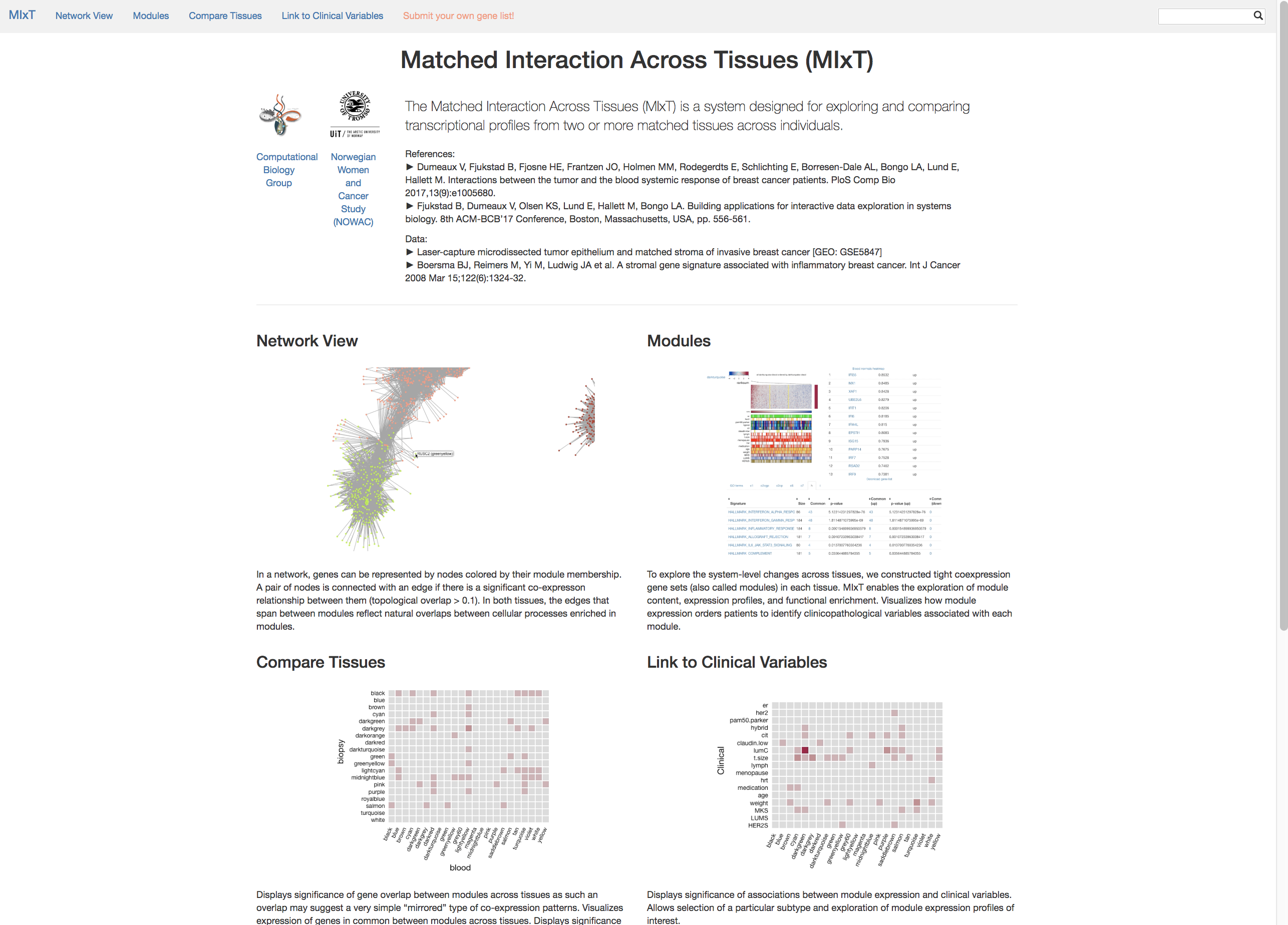
MIxT: system designed for exploring and comparing transcriptional profiles from two or more matched tissues across individuals.
Relevant websites:
Tumor-blood interactions in breast cancer patients
Tumor epithelium-stroma interactions in breast cancer
Unraveling the functionality of microbiomes with novel computational and single-cell profiling approaches
Although there has been an explosion in methods and applications of single-cell genomic profiling in mammalian systems, single-cell profiling approaches for measuring DNA or RNA in individual microbes remain in their infancy. 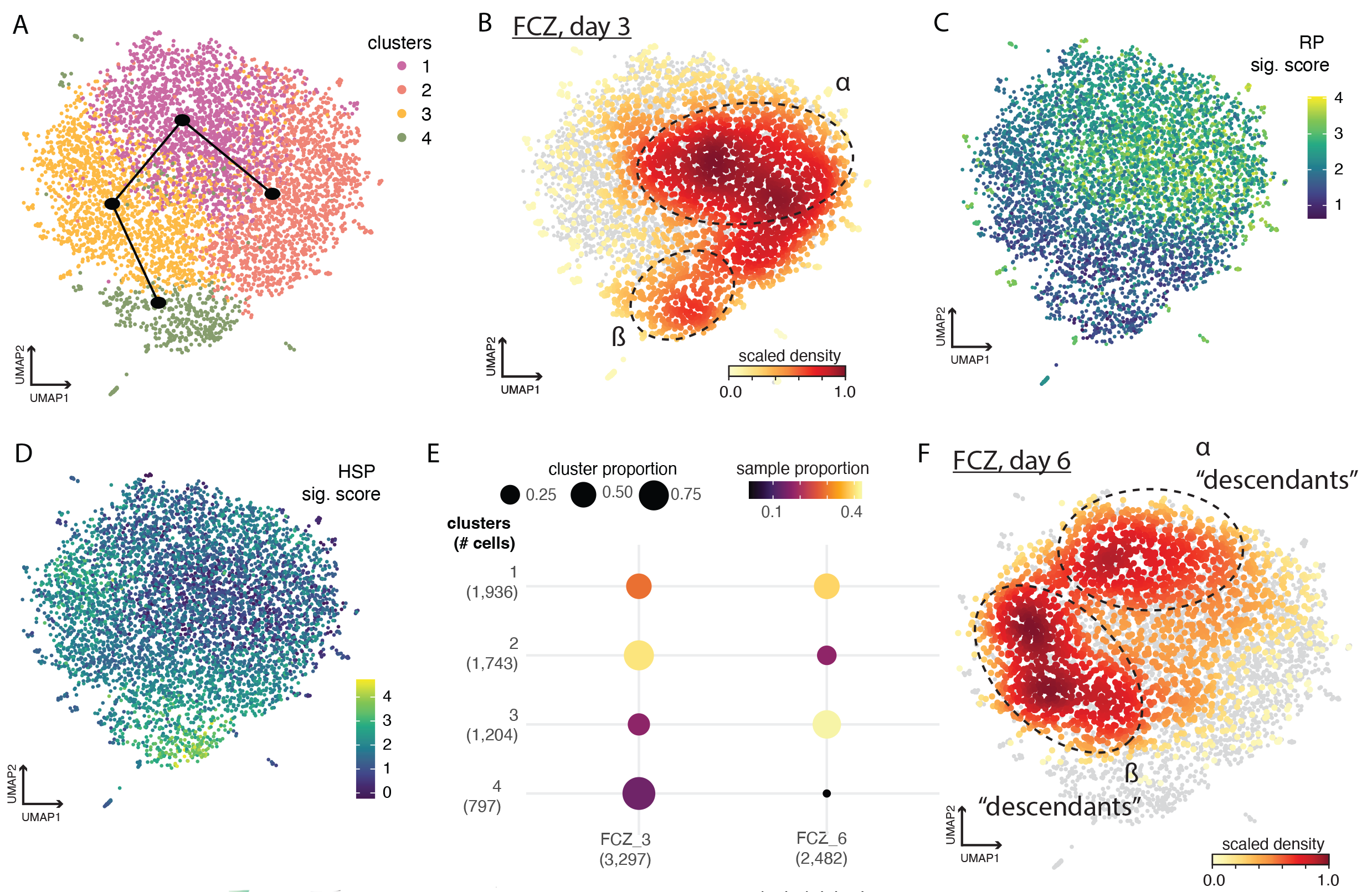 Nevertheless, the field of microbiology recognizes single-cell approaches as a promising tool, for example, to characterize heterogeneity in functional responses to different conditions or challenges. The difficulty of microbial single-cell approaches is at least partly due to their physical and molecular properties. Our recent work (Elife 2023) was the first to tailor a nanolitre-droplet assay to profile individual transcriptomes of the fungal pathogen Candida albicans. We used this device to explore phenotypic heterogeneity in a laboratory strain of this fungus. Our study provided deeper insight into how genetically identical individuals have different responses to the same stresses that ultimately enable some individuals to survive and others to die. The heterogeneity observed in monocultures will likely be greatly magnified in complex microbial environments such as the gut microbiome.
Nevertheless, the field of microbiology recognizes single-cell approaches as a promising tool, for example, to characterize heterogeneity in functional responses to different conditions or challenges. The difficulty of microbial single-cell approaches is at least partly due to their physical and molecular properties. Our recent work (Elife 2023) was the first to tailor a nanolitre-droplet assay to profile individual transcriptomes of the fungal pathogen Candida albicans. We used this device to explore phenotypic heterogeneity in a laboratory strain of this fungus. Our study provided deeper insight into how genetically identical individuals have different responses to the same stresses that ultimately enable some individuals to survive and others to die. The heterogeneity observed in monocultures will likely be greatly magnified in complex microbial environments such as the gut microbiome.
Host-adapted microbial communities, commonly referred to as microbiomes, consist of diverse microorganisms including commensal, symbiotic, and pathogenic species occupying specific anatomical niches. Traditionally, community compositions are quantified as vectors of relative abundances at various taxonomic levels. Studies have found that host-adapted microbiomes are variable across individuals, yet certain configurations are observed more frequently across the global population than what we would expect by chance. Multiple factors, ranging from environmental pressures to intrinsic microbe-microbe interactions, could explain the existence of these preferred configurations.
In humans, significant efforts have been dedicated to defining dominant microbial configurations (MCs) within various microbiomes, including those of the vagina, skin, oral cavity, and gut. In the gut, MCs, also called enterotypes, have been associated with dietary, and various metabolic or immunological markers. Despite these advancements, the functional roles and the underlying mechanisms contributing to the stability and resilience of these MCs remain poorly understood.
Our research is developing novel computational approaches to identify and characterize specific MCs. We apply deep learning to analyze a large compendium of healthy adult gut microbiomes, capturing non-linear trends between features, such as taxa or pathway abundances. These efforts aim to refine the definition and understanding of both compositional and functional MCs. As we delve deeper into the intricacies of MCs, the opportunities to modulate them for varied applications in biomedicine, environment, and synthetic biology will expand.
Relevant recent papers:
Candida albicans exhibits heterogeneous and adaptive cytoprotective responses to anti-fungal compounds
Deep learning reveals functional archetypes in the adult human gut microbiome that underlie interindividual variability and confound disease signals
Completed projects
Development of RNA profiles of blood cells as a tool to study individual’s exposure and disease
We have had a long-standing interest in the development of RNA-based biomarker signatures in blood, which can be used to inform on exposure and health status. As a major defense and transport system, blood cells can adjust expression of their genes in response to various clinical, biochemical, and pathological conditions. To develop such surrogate signatures, we first defined robust laboratory methodologies for RNA profiling of blood cells (Dumeaux et al, 2008 Biomarkers in Medicine), and investigated “normal” inter-individual variation in healthy individuals (Dumeaux et al, 2010 PloS Genetics). These studies served as stepping-stones to several projects that investigated how the signatures can be used to detect systemic molecular processes deregulated in response to defined exposure and health status (>5 publications, 200+ citations). These manuscripts highlight specific behavioral programs, such as metabolism or signaling, deregulated in the individual’s blood cells that are biological and/or pathological responses to a given condition in the general population (eg exposure to organic pollutants) or in a diseased population (eg radiation-induced fibrosis in breast cancer survivors). These blood-based signatures can also be used to develop new ways to diagnose the disease. For example, our blood-based diagnostic test for breast cancer may reduce false positive interpretations of suspicious mammographic results (3 publications including Dumeaux et al, 2015 IJC; one submitted patent).
Relevant recent work:
Peripheral blood cells inform on the presence of breast cancer: a population-based case-control study
Deciphering normal blood gene expression variation--The NOWAC postgenome study
Using blood gene signatures for assessing effects of exposure to perfluoroalkyl acids (PFAAs) in humans: the NOWAC postgenome study
Sex hormones and gene expression signatures in peripheral blood from postmenopausal women - the NOWAC postgenome study
Development of novel bioinformatics approaches and softwares
We develop approaches that balance and inter-connect quantitative and biological knowledge. Many acknowledge that no field has generated higher expectations, deeper frustrations, and more “translation anxiety” than advances in human genomics. Early on, we among others have highlighted the role of rigorous epidemiological and statistical approaches in improving the prospect of genomics and big data in personalized medicine. Our approach, termed “Systems Epidemiology” (Lund & Dumeaux, 2008 CEBP), proposes to integrate human -omics data with measurements from observational epidemiologic studies to better characterize the diverse range of factors influencing complex diseases, and help infer causation and support evidence-based research (Lund & Dumeaux, 2010 Int J Epi). In line with these concepts, we supported the development of a large biobank within the Norwegian Women and Cancer Study (Dumeaux et al, 2008 BCR).
Also, critical to these efforts is the development of computational methodologies that support the integration and interpretation of these complex “real-life” data. Specifically, we have developed novel methodologies for the sensitive detection of low amplitude changes in blood profiles across healthy individuals (developed in PloS Genetics 2010), for identifying genes, pathways, and processes that co-vary and interact across tissues and environments, for predicting activation of molecular pathways in a single-patient manner satisfying clinical practice constraints associated with personalized medicine, as well as other methodologies within collaborative manuscripts (Huttenhower et al, 2009 Genome Research; Barutcuoglu Z et al, 2009 Bioinformatics; Bettauer et al, 2022 Microbiology Spectrum)
Relevant recent papers:
Interactions between the tumor and the blood systemic response of breast cancer patients
Detecting gene signature activation in breast cancer in an absolute, single-patient manner.
Building applications for interactive data exploration in systems biology.
A Deep Learning Approach to Capture the Essence of Candida albicans Morphologies.
Relevant recent software packages:
MIxT: system designed for exploring and comparing transcriptional profiles from two or more matched tissues across individuals.
Candescence
Relevant websites:
Tumor-blood interactions in breast cancer patients
Tumor epithelium-stroma interactions in breast cancer
Contribution of the brain-gut-microbiota axis in health
Each human gut harbors communities of trillions of microbes called microbiota. Collectively, these microbes have about 150 to 300 times as many genes (the microbiome) as the human genome. This vast army of microbes (and genes) is now recognized as a vital component in furnishing and maintaining human health. They deliver minerals and vitamins, help digest our food, can affect our mood and immune system. Recent research shows that the central nervous system and the digestive tract maintain a two-way line of communication, the “brain-gut-microbiome” axis.  By secreting gas, chemical or even neurotransmitters, the gut microbiota can affect defined brain areas related to eg. stress/anxiety, satiation, sleep, memory, and cognitive functioning. Although these results are promising to explain some microbiome-derived health benefits related to environmental stimuli, studies are challenged by the fact that the human microbiome is complex and varied. We analyzed profiles of the gut microbiota and brain activity before and after an intervention, a design that should transcends conventional approaches capturing only a single screenshot of the microbiome.
By secreting gas, chemical or even neurotransmitters, the gut microbiota can affect defined brain areas related to eg. stress/anxiety, satiation, sleep, memory, and cognitive functioning. Although these results are promising to explain some microbiome-derived health benefits related to environmental stimuli, studies are challenged by the fact that the human microbiome is complex and varied. We analyzed profiles of the gut microbiota and brain activity before and after an intervention, a design that should transcends conventional approaches capturing only a single screenshot of the microbiome.








