Abstract
Postmenopausal hormone therapy (HT) influences endogenous hormone concentrations and increases the risk of breast cancer. Gene expression profiling may reveal the mechanisms behind this relationship.Our objective was to explore potential associations between sex hormones and gene expression in whole blood from a population-based, random sample of postmenopausal women
Gene expression, as measured by the Applied Biosystems microarray platform, was compared between hormone therapy (HT) users and non-users and between high and low hormone plasma concentrations using both gene-wise analysis and gene set analysis. Gene sets found to be associated with HT use were further analysed for enrichment in functional clusters and network predictions. The gene expression matrix included 285 samples and 16185 probes and was adjusted for significant technical variables.
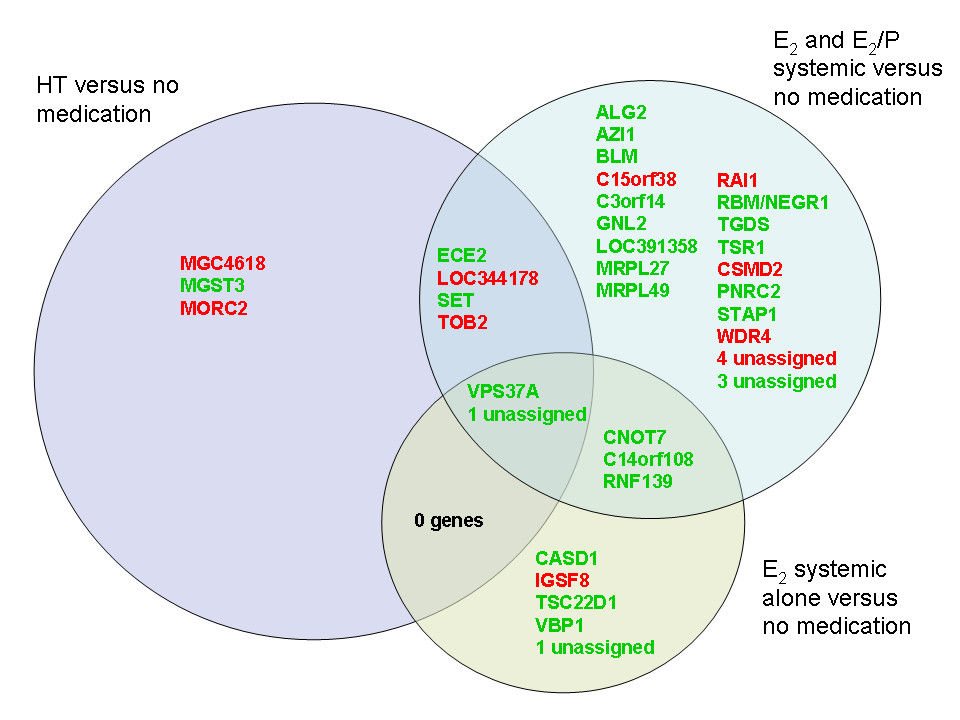
Gene-wise analysis revealed several genes significantly associated with different types of HT use. The functional cluster analyses provided limited information on these genes. Gene set analysis revealed 22 gene sets that were enriched between high and low estradiol concentration (HT-users excluded). Among these were seven oestrogen related gene sets, including our gene list associated with systemic estradiol use, which thereby represents a novel oestrogen signature. Seven gene sets were related to immune response. Among the 15 gene sets enriched for progesterone, 11 overlapped with estradiol. No significant gene expression patterns were found for testosterone, follicle stimulating hormone (FSH) or sex hormone binding globulin (SHBG).
Distinct gene expression patterns associated with sex hormones are detectable in a random group of postmenopausal women, as demonstrated by the finding of a novel oestrogen signature.