Abstract
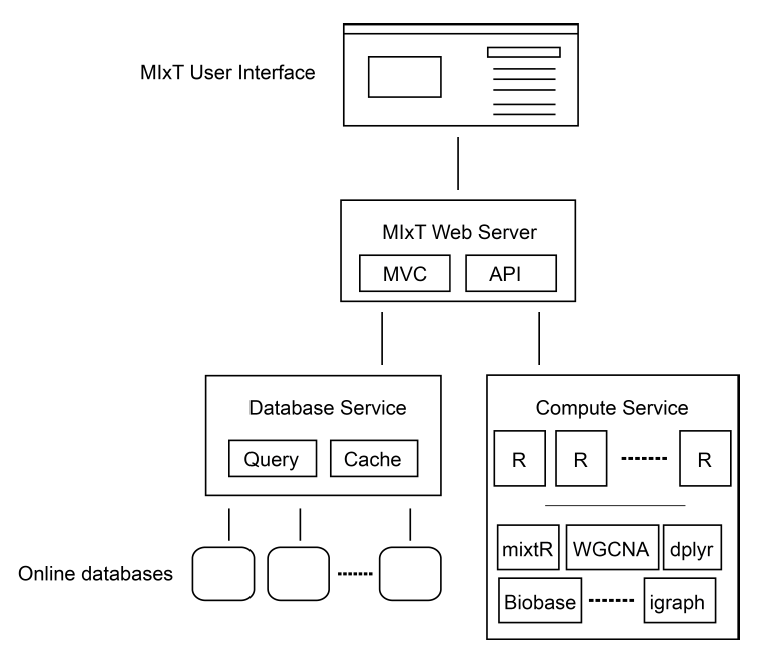
The significant increase in the rate of data generation by the systems biology community creates a need for interactive exploration tools to explore the resultant datasets. Such tools need to combine advanced statistical analyses, prior knowledge from biological databases, and interactive visualizations with intuitive user interfaces. Each specific research question potentially requires a specialized user interface and visualization methods. Although some features are application-specific, the underlying components of the data analysis tool can be shared and reused. Our approach for developing data exploration tools in systems biology builds on the microservice architecture that separates an application into smaller components which can communicate using language-agnostic protocols. We show that this design is well suited for bioinformatics applications where different tools written in different languages by different research groups is the norm. Packaging each service in a software container enables re-use and sharing of key components between applications, reducing development, deployment, and maintenance time. We demonstrate the viability of our approach through a web application, entitled MIxT blood-tumor, for exploring and comparing transcriptional profiles from blood and tumor samples in breast cancer patients. The application integrates advanced statistical software, up-to-date information from biological databases, and modern data visualization libraries.