Abstract
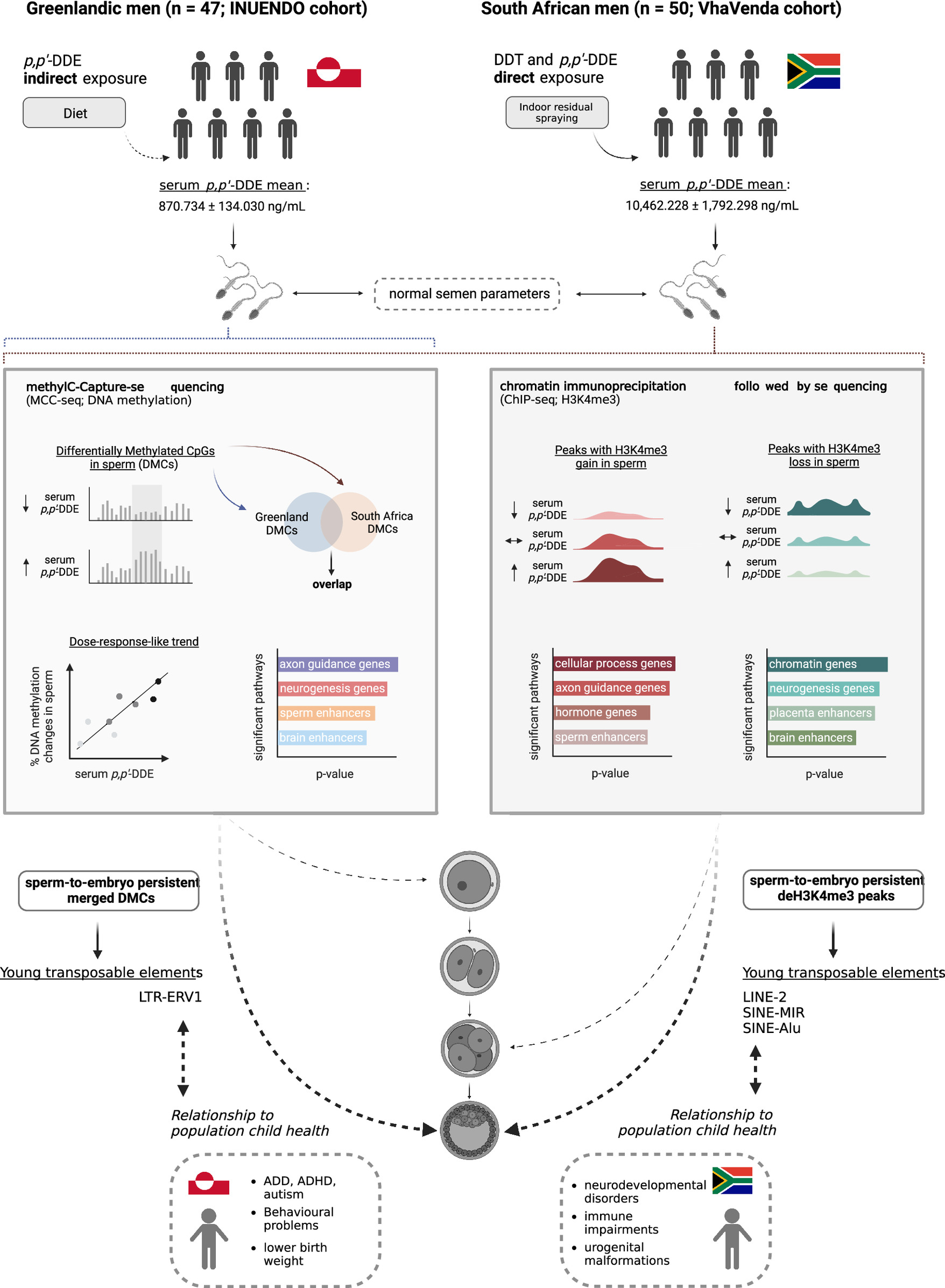
Background: The organochlorine dichlorodiphenyltrichloroethane (DDT) is banned worldwide owing to its negative health effects. It is exceptionally used as an insecticide for malaria control. Exposure occurs in regions where DDT is applied, as well as in the Arctic, where its endocrine disrupting metabolite, p,p’-dichlorodiphenyldichloroethylene (p,p’-DDE) accumulates in marine mammals and fish. DDT and p,p’-DDE exposures are linked to birth defects, infertility, cancer, and neurodevelopmental delays. Of particular concern is the potential of DDT use to impact the health of generations to come via the heritable sperm epigenome.
Objective: The objective of this study was to assess the sperm epigenome in relation to p,p’-DDE serum levels between geographically diverse populations.
Methods: In the Limpopo Province of South Africa, we recruited 247 VhaVenda South African men and selected 50 paired blood serum and semen samples, and 47 Greenlandic Inuit blood and semen paired samples were selected from a total of 193 samples from the biobank of the INUENDO cohort, an EU Fifth Framework Programme Research and Development project. Sample selection was based on obtaining a range of p,p’-DDE serum levels (mean = 870.734 +/- 134.030 ng/mL). We assessed the sperm epigenome in relation to serum p,p’-DDE levels using MethylC-Capture-sequencing (MCC-seq) and chromatin immunoprecipitation followed by sequencing (ChIP-seq). We identified genomic regions with altered DNA methylation (DNAme) and differential enrichment of histone H3 lysine 4 trimethylation (H3K4me3) in sperm.
Results: Differences in DNAme and H3K4me3 enrichment were identified at transposable elements and regulatory regions involved in fertility, disease, development, and neurofunction. A subset of regions with sperm DNAme and H3K4me3 that differed between exposure groups was predicted to persist in the preimplantation embryo and to be associated with embryonic gene expression.
Discussion: TThese findings suggest that DDT and p,p’–DDE exposure impacts the sperm epigenome in a dose–response-like manner and may negatively impact the health of future generations through epigenetic mechanisms. Confounding factors, such as other environmental exposures, genetic diversity, and selection bias, cannot be ruled out.