Abstract
Background Understanding the functional diversity of the gut microbiome is critical for elucidating its roles in human health and disease. While traditional approaches focus on taxonomic composition, functional configurations of the microbiome remain understudied. This study introduces a deep-learning framework combined with archetypal analysis to identify and characterize functional archetypes in the adult human gut microbiome. This approach aims to provide insights into interindividual variability, function-driven microbiome stability, and the potential confounding role of functional diversity in disease-associated microbial signatures.
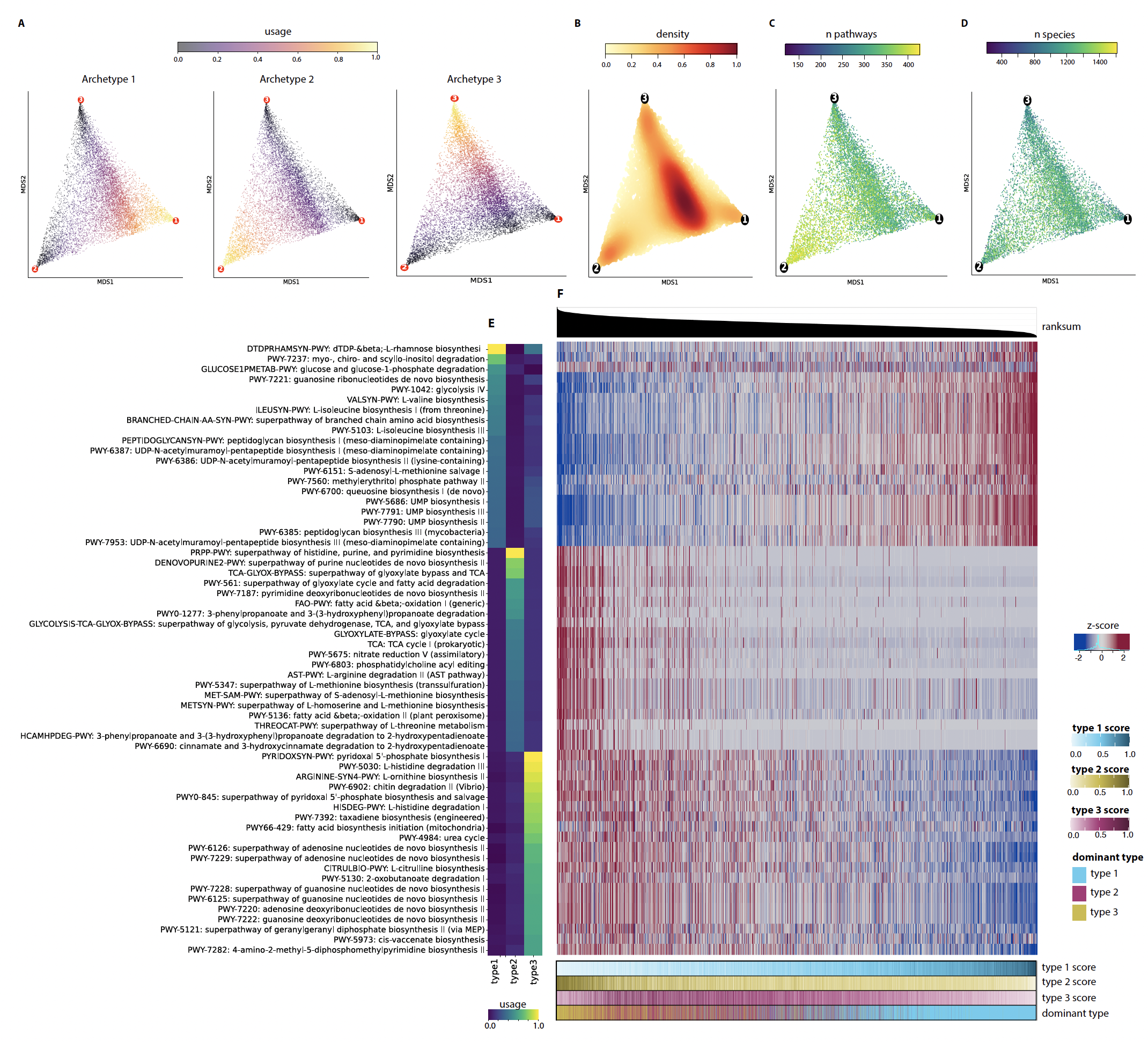
Results Analyzing 9838 whole-genome metagenomic samples from healthy adults across 29 countries, we identified three distinct functional archetypes that define the boundaries of the gut microbiome’s functional space. Each archetype is characterized by unique metabolic potentials: Archetype 1 is enriched in sugar metabolism, branched-chain amino acid biosynthesis, and cell wall synthesis; Archetype 2 is dominated by fatty acid metabolism and TCA cycle pathways; and Archetype 3 is defined by amino acid and nitrogen metabolism. While most gut microbiome communities are a blend of these archetypes, some align closely with a single archetype, potentially reflecting adaptation to host factors such as distinct dietary patterns. Proximity to these archetypes correlates with microbiome stability, with Archetype 2 representing the most resilient state, likely due to its metabolic flexibility and diversity.
Functional archetypes emerged as a potential confounder in disease-associated microbial signatures, including in type-2 diabetes, colorectal cancer, and inflammatory bowel disease (IBD). In IBD, archetype-specific shifts were observed: Archetype 1-dominant samples exhibited increased carbohydrate metabolism, while Archetype 3-dominant samples showed enrichment in inflammatory pathways. These findings highlight the potential for archetype-specific functional changes to inform microbiome-targeted interventions.
Conclusions The identified functional archetypes provide a robust framework for addressing interindividual variability and potential confounding in gut microbiome-based disease studies. By incorporating archetypes as potential confounders or stratification factors, researchers can reduce variability, uncover novel pathways, and improve the precision of microbiome-targeted interventions. The deep-learning framework can be applied to other host-associated microbial ecosystems, providing new insights into microbial functional dynamics and their implications for the host’s health.