Abstract
Whole blood is likely to become the prime tissue to detect biomarkers using gene expression analyses. In this study, we assessed whether whole-blood or globin-reduced RNA provides the most robust and sensitive results to detect small gene expression changes, for example, in response to hormone therapy (HT) exposure.
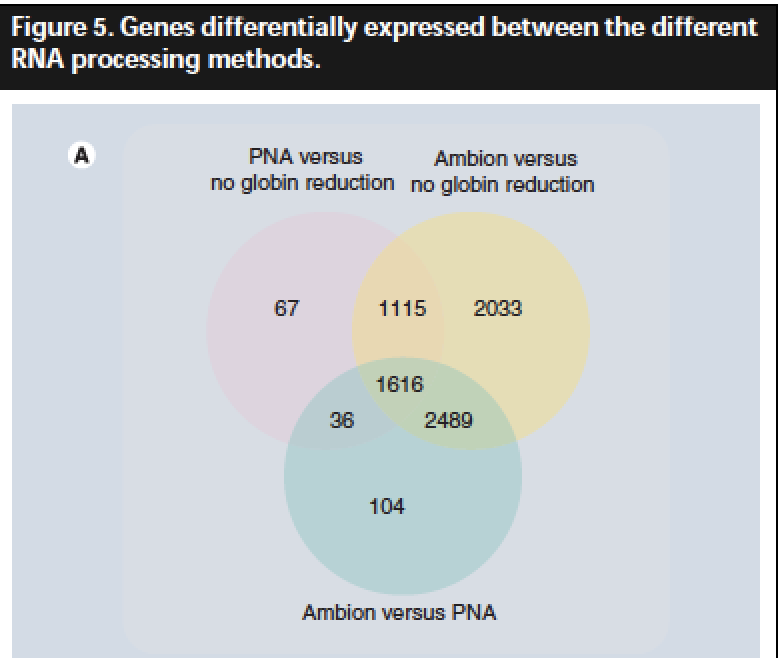
Each sample (n = 12) was processed according to three different protocols: no globin reduction, globin reduction using peptide nucleic acids (PNAs) and globin reduction using magnetic beads in the GlobinClear kit from Ambion.
Both globin reduction approaches using Ambion kit and PNAs were efficient at reducing globin RNA. However, globin reduction using Ambion kit also lowered the remaining cRNA. Samples processed with PNAs gave an intermediary profile closest to the no globin reduction group, with a slight increase in sensitivity of transcript detection and decrease in variability, but a loss of reproducibility.
Globin RNA processing method in blood transcriptome analyses should be optimized for each microarray platform to be used. In our study, globin reduction was not found to be beneficial using the Applied Biosystems microarray system (AB1700).