Abstract
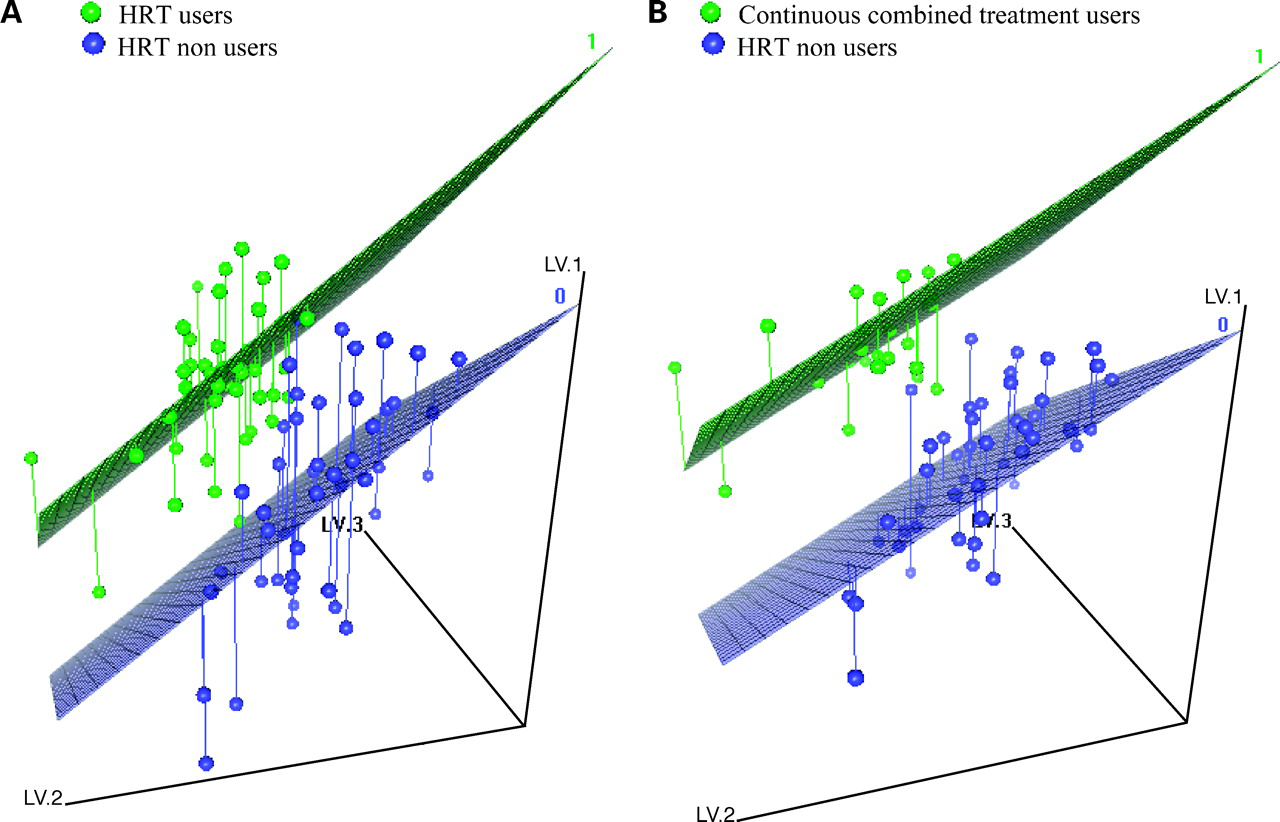
The American Women’s Health Initiative study published in July 2002 caused considerable concern among hormone replacement therapy (HRT) users and prescribers in many countries. This study is an exploratory research comparing the genome-wide expression profile in whole-blood samples according to HRT use. Within the Norwegian Women and Cancer study, 100 postmenopausal women (50 HRT users and 50 non-HRT users) born between 1943 and 1949 with normal to high body mass index and no other medication use were selected. After total RNA extraction, amplification, and labeling, the samples were hybridized together with a common reference (Universal human reference RNA, Stratagen) to Agilent Human 1A oligoarrays (G4110b, Agilent Technologies) containing 20,173 unique genes. Differentially expressed genes were used to build a classifier using the nearest shrunken centroid method (PAM). Then, we tested the significant changes in single genes by different methods like t test, Significance Analysis of Microarrays, and Bayesian ANOVA analysis. Results did not reveal any distinct gene list which predicted accurately HRT exposure (error rate, 0.40). Classifier performance slightly improved (error rate, 0.26) including only women who were using continuous combined HRT treatment. According to the small amplitude of expression alterations observed in whole blood, more quantitative technique and larger sample sizes will be needed to be able to investigate whether significant single genes are differentially expressed in HRT versus non-HRT users. Taken cautiously, significant enrichments in biological process of genes with small changes after HRT use were observed (e.g., receptor and transporter activities, immune response, frizzled signaling pathway, actin filament organization, and glycogen metabolism).